Step by Step: Identifying High-Risk Tumour Regions in Sarcoma
Introduction
In my capstone project, we are interested in using deep learning to create saliency maps to inform radio oncologists of regions for potential focal dose escalation to prevent metastasis of soft tissue sarcoma (STS) - a rare yet aggressive cancer arising in connective tissues.
One of the challenges in treatment of STS, particularly Extremity Soft-Tissue Sarcoma (ESTS), is the precision of targeted radiation therapy. While AI has made significant strides in automatic tumour segmentation, its clinical implications remain uncertain since segmentation accuracy focuses on the gross tumour volume rather than the much larger planning target volume accounting for variations due to tissue swelling and the spread of disease into adjacent tissues, among other factors.
Therefore, we are looking into the identification of high-risk tumour subregions within sarcomas. These areas, potentially more resistant to radiation, are being pinpointed with the aid of deep learning models. Our objective is to improve radiation therapy by offering a more personalized approach, targeting specific tumour areas for increased dosage while safeguarding the surrounding healthy tissues.
Another intriguing future aspect of our project is the correlation of high-risk areas identified by ai with histopathological results. We are exploring whether these regions correspond to a higher concentration of cancer stem cells or show less tumour necrosis following pre-operative radiation. This correlation is crucial in understanding the biological underpinnings of these high-risk areas.
MRI pre-processing
We are using an open source dataset of STS images from TCIA. It consists of 51 patients with non-registered versions of T1 & T2 magnetic resonance (MR) images and registered versions of T1, T2, PET, and CT scans.
For the initial part of our project, we are interested in using only the MR images since they provide the best contrast for soft tissue imaging. To complete the pre-processing of these images, we: 1) apply n4 bias correction to the non-registered T1 and T2 images and 2) register the bias-corrected images by converting the binary radio therapy contour masks to nifti volumes, registering these structures using simpleITK, and then applying the calculated transformation to the bias corrected image itself.
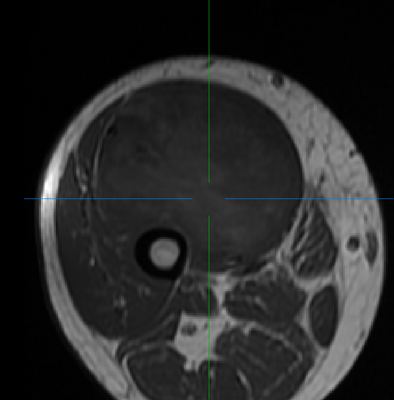
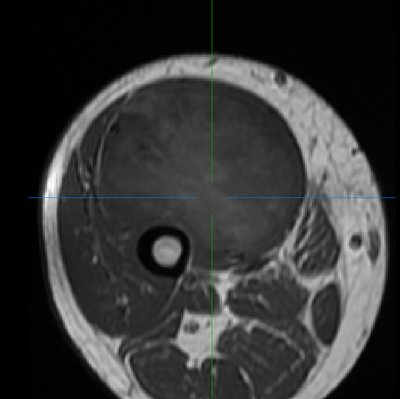
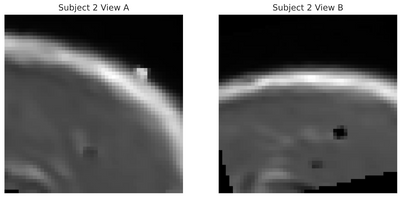
(Left/Top) T1 registered image without bias correction. (Right/Bottom) T1 image post bias correction and registration. Higher contrast and greater uniformity is apparent.
When we wrap up this project in April 2024, the entire codebase will be available to examine exactly how these steps are performed. In the meantime, reach out if you have any questions.
3D simCLR
Note: This work is currently in progressOur project employs the simCLR (Simple Framework for Contrastive Learning of Visual Representations) framework, particularly beneficial given our dataset's limitation in size and high variability. This self-supervised approach aims to learn an embedding space where similar sample pairs are close while dissimilar ones are distant.
We implemented this using PyTorch Lightning for efficient management of the training loop and integrated metric logging with Weights & Biases. Image augmentation, a key component in simCLR, was performed using TorchIO. Unlike supervised learning, where certain augmentations might degrade performance, contrastive learning benefits from these augmentations, encouraging the learning of more generalizable features.
In adapting to our 3D volumetric data, we evolved from the ResNet-50 to a 3D ResNeXt-50 architecture. This model's 'split-transform-merge' strategy introduces a dimension called cardinality, or the number of parallel filter sets within a layer, which has been shown to more effectively improve model performance compared to increasing depth or width.
To manage the large size of our input volumes, we used a sliding kernel technique, breaking down images into smaller, overlapping patches. This allowed for larger batch sizes, essential for simCLR’s effectiveness, especially with our limited dataset and constrained training epochs. These larger batch sizes provide a diverse set of negative samples, crucial for the effective convergence of the contrastive loss function.